By Ma Chunlei

“Sour, sweet, bitter, salty and umami” are widely recognized as the five perceptible tastes, of which “salty” is a necessary taste feeling. Currently, the per capita daily Na+ intake is more than twice the recommended intake of the World Health Organization (WHO; less than 2 g/d Na+ or 5 g/d NaCl). Excessive consumption of sodium salt is one of the important inducers of cardiovascular and cerebrovascular diseases. At present, various countries have proposed different solutions to reduce the NaCl content in foods without changing the original salty taste, such as replacement with other metal salts, addition of flavor enhancers, and improvement of the taste perception of NaCl through taste contrast.
Salty peptides are mainly composed of L-type amino acids, and are mainly composed of two or more amino acid peptide bonds, while polypeptides are composed of two or more amino acid peptide bonds. The number of salty peptides reported in the study is small, mainly dipeptides. These dipeptides have a variety of taste characteristics such as sour taste, fresh taste and salty taste. The salty peptide ionizes the cation, enters the cell through the sodium ion channel on the cell membrane, and then polarizes the calcium ion. When the internal part of the cell is positively charged, a small current will be formed, and then its conduction medium will be released. The primary input to the neuron will send a "salty" taste to the brain, which will be salty. See Figure 1 for saltiness transmission mechanism.
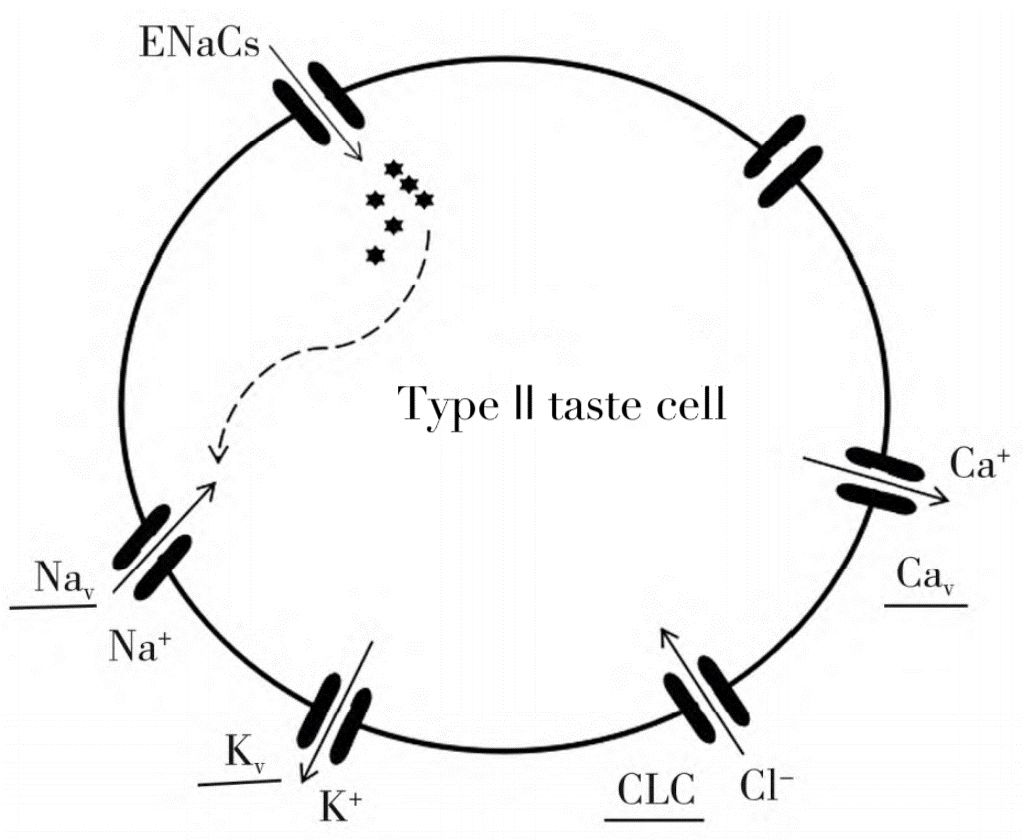
Fig. 1. The saltiness transmission mechanism (Zhang et al. 2018).
Yeast extract (YE) is a natural and environment-friendly salty and umami flavor enhancer rich in amino acids, 5′-nucleotides and peptides, which is widely used in various condiments and processed foods. Inguglia et al clearly proposed that the use of flavor enhancers can increase salty taste without increasing salt; Zheng et al isolated, purified and identified five peptides with salty-enhancing characteristics from YE. However, the current lack of understanding of the structure of the receptor that causes saltiness makes the identification or screening of salt- enhancing peptides dependent on a great deal of work. On the contrary, the 3D structure of umami receptor T1R1/T1R3 has been elucidated by computational modeling, and the research system of umami peptides using molecular docking technology is relatively mature. In addition, the recognition of salty taste is closely related to salty taste receptors, and there is little research on salty peptides and salty enhancing peptides at the molecular level.
Therefore, the separation, purification and identification of salty peptides in YE and the salty mechanism were studied as follows:
1. Identification and characterization of peptides from YE
Many studies have been carried out to determine the salty enhancing peptides of YE (KU012) through sensory evaluation experiments, as shown in Fig. 2-A. The salty taste of the peptide less than 1 kDa was the most obvious, although that of the peptide with the MW between 3 kDa and 5 kDa was stronger, and weaker. Fig. 2-B shows that the peptide fractions with an MW less than 1 kDa obtained by ultrafiltration were separated by further gel filtration and separated into five components: G-1, G-2, G-3, G-4, and G-5. Sensory evaluation showed that the highest salty flavor of the G-1 component was 4 points (recovery rate was 0.9%), and the other components had no obvious salty flavor.
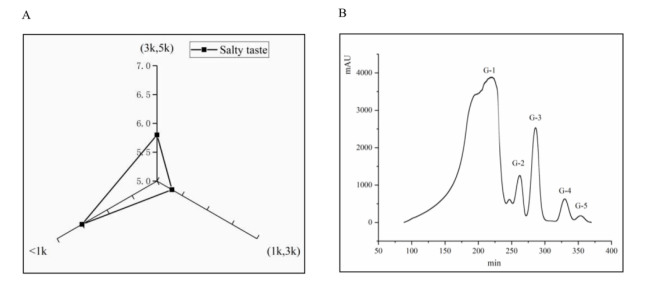
Fig. 2. Salty taste of three MW peptides in YE (A) and chromatograms of (B)
GFC.
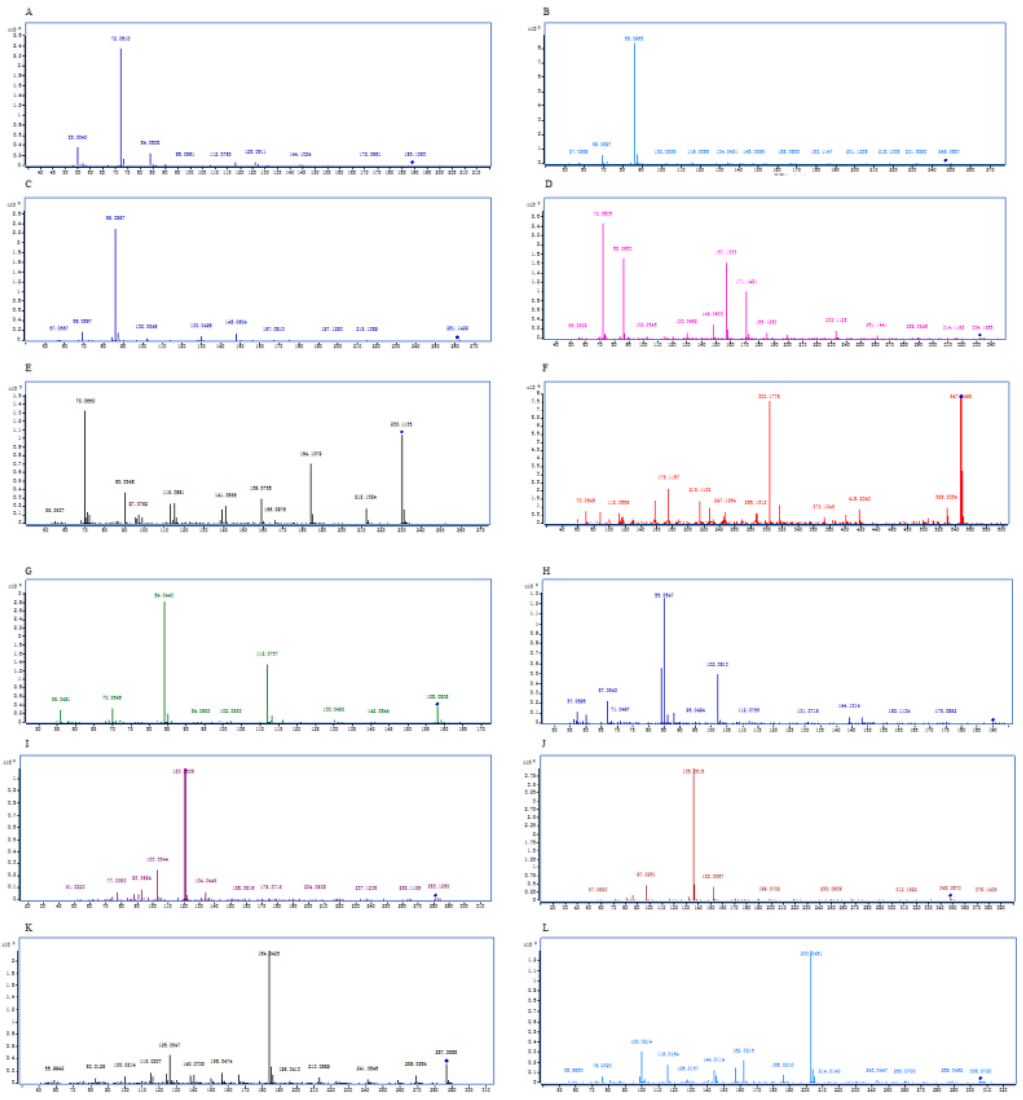
Fig. 3. Tandem mass spectra of UPLC–ESI-Q-TOF MS/MS. Peptides are (A) AV; (B) NM; (C) NE; (D) SPE; (E) PN; (F) KSAEN; (G) AT; (H) GD; (I) CY; (J) NSE; (K/L)
IR/LR; M CVS.
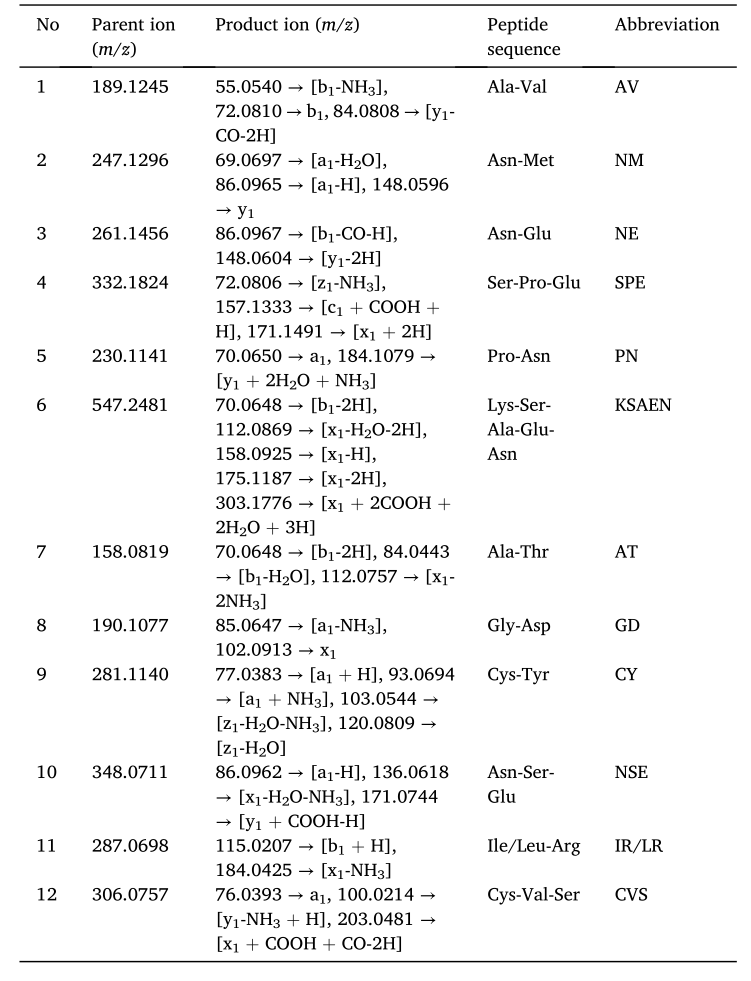
The amino acid sequence of the fragment was determined and predicted by UPLC–ESI-Q-TOF MS/MS, as shown in Fig. 3. As shown in Table 1 and Fig. 3, thirteen possible salty enhancing peptides in YE (KU012) were identified by UPLC–ESI-Q-TOF MS/MS: AV, NM, NE, SPE, PN, KSAEN, AT, GD, CY, NSE, IR/LR and CVS.
Table 1 Fragment ions and proposed peptide sequence.
2. Construction and evaluation of salty receptor TMC4
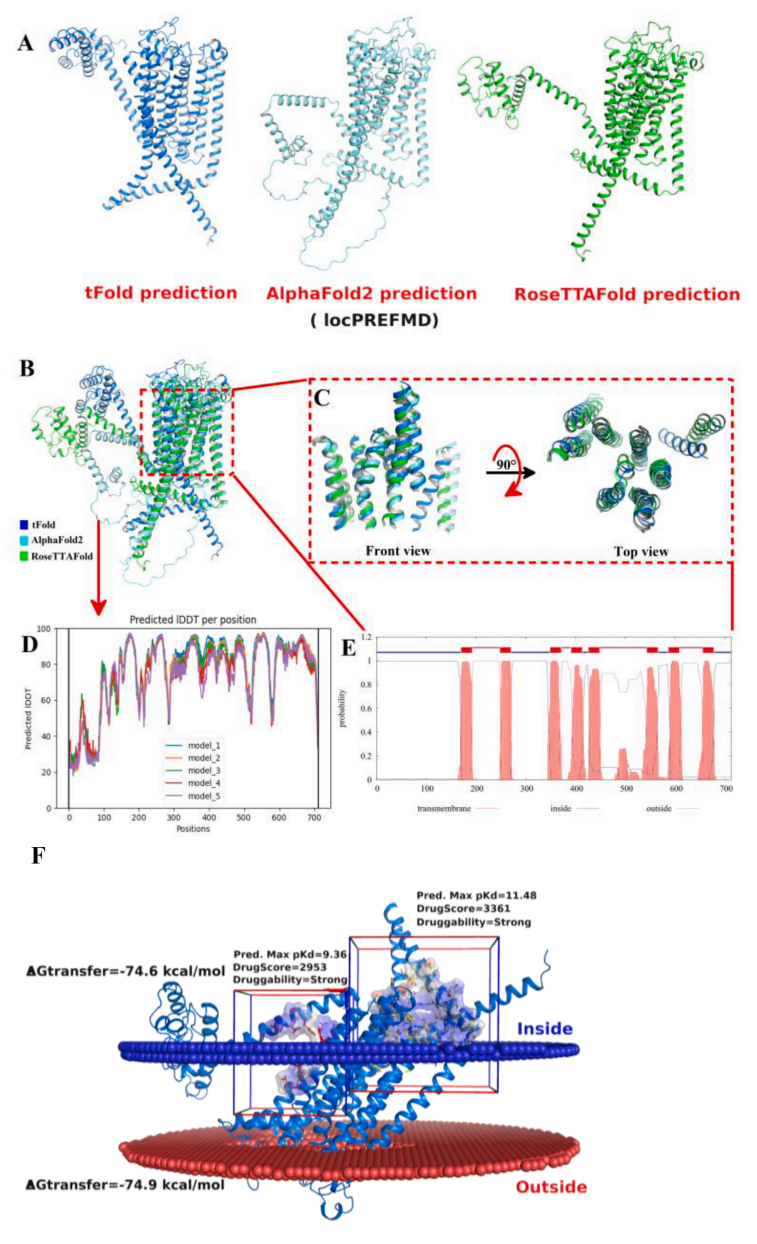
Fig. 4. Construction and evaluation of 3D structure of TMC4 with eight transmembrane domains. (A) 3D structure of TMC4 build via tFold, AlphaFold2 and RoseTTAFold, and refined with locPREFMD. (B) Structural comparison of the transmembrane regions of TMC4 protein models. (C) Partial front- and top view of TMC4. (D) Predicted IDDT per position of Alphafold2 and (E) Predicted of transmembrane regions with the TMHMM server. (F) Transmembrane state and potential allosteric sites of TMC4 receptor protein.
TMC4 is a voltage-dependent chloride channel and membrane protein with eight transmembrane domains that plays a key role in human perception of saltiness. As shown in Fig. 4, the final 3D structure of TMC4 with eight transmembrane domains was predicted using tFold, AlphaFold2 and RoseTTAFold and refined with locPREFMD, respectively. (Fig. 4-A).
From the perspective of the three-dimensional structure, the threedimensional structures of eight transmembrane domains constructed by tFold, Alphafold2 and RoseTTAFold were generally the same, while the structures of 186 amino acid residues in front of the N-terminus were very different (Fig. 4-BC). The average predicted IDDT of these transmembrane regions is higher than 85%, which indicates that the structure of this region is reliable (Fig. 4-DE).
The transmembrane state, and potential allosteric sites of TMC4 were further explored based on the optimal model, and the results are shown in Fig. 3-F. It is speculated that the salt enhancing peptide may activate TMC4 by binding to one of the pockets. Therefore, two pockets were used for molecular docking.
3. Recognition and mechanism of salty enhancing peptides
Six synthesized peptides, KSAEN, PN, CY, NSE, NE and SPE (purity >98%), were prepared at 1% (m/v). Fig. 5 shows a 2D diagram of the interaction of peptide segments in the docking mode of energy minimization in molecular docking with the receptor. Through the analysis of the binding sites and binding forces between the peptide and TMC4 receptor, it was found that there were mainly hydrogen bonding and hydrophobic interactions in the binding process. Thus, Table 2 lists the key amino acid residues around the activesites with hydrogen bonding forces when the salty-enhanced peptide interacts with TMC4. The types of amino acid residues affected by hydrogen bonding are few and concentrated in pocket 2, and pocket 4 is more abundant and dispersed than pocket 2. In addition, the docking results showed that Arg151, Thr148 and Tyr565 played a key role in the interaction between the peptide and receptor (Pocket 2) mainly through hydrogen bond forces and salt bridges. In pocket 4, Arg424, Arg330, Tyr677 and Arg580 are also important binding sites. In pocket 2, Trp147, Ile152, Lys568 and Phe572 are important binding sites, while in pocket 4, there are also Leu684, Thr421, Lys685, Ala333 and other binding sites. Based on the analysis of binding modes and binding sites between these peptides and receptors, hydrogen bonds, hydrophobic interactions and electrostatic interactions are usually necessary factors for the interaction between receptors and ligands. Amino acid residues (Asn, Glu and Ser) can bind to the amino acid residues of TMC4 through interaction. Theoretically, the four salty-enhancing peptides have good interaction with the conformation of TMC4 and can bind to pocket 2 and pocket 4.
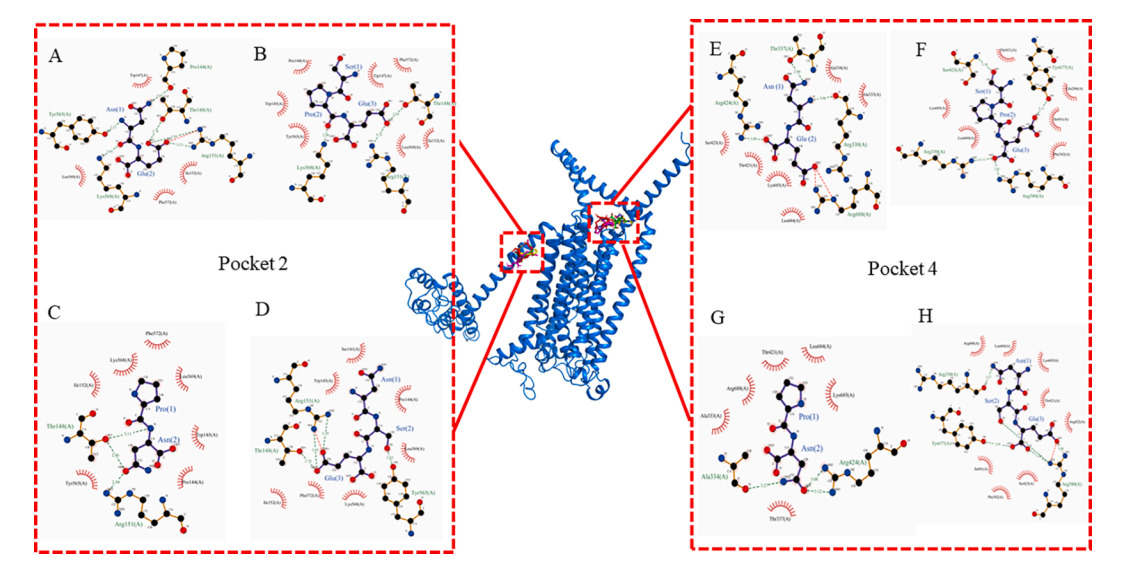
Fig. 5. 3D and 2D structural diagram of docking between four salty-enhancing peptides and TMC4 receptor protein. Pocket 2 is on the left and pocket 4 on the right. Peptide segments are distinguished according to different colors, in which green is NE (A and E); red is SPE (B and F); yellow is PN (C and G); magenta is NSE (D and H). Ligand bonds are purple, non-ligand bonds are orange, hydrogen bonds are olive green, salt bridges are red, disulfide bonds are gold, and hydrophobic bonds are brick red.
Table 2 The key amino acid residues around the active sites in salty enhancing peptidesand TMC4
pockets | TMC4 residues | NE | SPE | PN | NSE |
Pocket 2 | Tyr565 | Na1b | S1 | ||
Lys568 | N1 | P1 | |||
Arg151 | E2 | E1 | N1 | E2 | |
Thr148 | E1 | E1 | N2 | E1 | |
Pro144 | N1 | ||||
Pocket 4 | Thr337 | N1 | |||
Arg424 | E1 | N2 | |||
Arg330 | N1 | E1 | N1 | ||
Arg668 | |||||
Ser423 | S1 | ||||
Arg580 | E1 | S1E2 | |||
Tyr667 | E1 | E1 | |||
Ala334 | N1 |
Note: a Affected amino acid; b Number of hydrogen bond force.
4. Conclusion
Human transmembrane channel-like 4 (TMC4) was constructed and evaluated by computer-based methods, and salt-enhancing peptides were identified based on its allosteric sites. PN, NSE, NE and SPE were further determined to be salty enhancing peptides, and their taste mechanism was investigated. It suggested that YE can be used as a raw material for the separation and purification of salty-enhancing peptides. The separation and purification of salty-enhancing peptides and the verification of the mechanism mean that YE not only has good flavor characteristics, but also clarifies its nutritional value, which is conducive to better social influence and utility. It can provide ideas and methods for the research and development of low-sodium products and new salty-enhancing peptides, and play a more important role in food flavor and nutrition in the future.
Reference:Shen DY, Pan F, Yang ZC, Song HL, Zou TT, Xiong J, Li K, Li P, Hu N, Xue DD. Identification of novel saltiness-enhancing peptides from yeast extract and their mechanism of action for transmembrane channel-like 4 (TMC4) protein through experimental and integrated computational modeling. Food Chem. 2022 Sep 15;388:132993.
About Angel Yeast
Founded in 1986, Angel Yeast Co., Ltd specializes in the production of yeast and yeast derivatives. Its product range includes baker's yeast and ingredients, Chinese dim sum and seasoning, savory yeast extract, human health, animal nutrition, plant nutrition, distilled spirits and biofuels, microbial nutrition and enzymes. At present, Angel Yeast has 11 international advanced production bases in China, Egypt and Russia, and provides products and services for more than 150 countries and regions globally.
About Angel Yeast Extract-Savoury:
Angel YE (yeast extract) made from edible yeast, by degradation the protein and nucleic acid in the yeast cells into nutritional seasonings with the application of modern biotechnology, has the advantages of increasing the fresh flavor, reducing salt, balancing the odor, strong tolerance and food properties, which promotes the global healthy operation of salt reduction and "clean label ".
Press Contact:
ANGEL YEAST CO.,LTD
Address: 168 Chengdong Avenue, Yichang, Hubei 443003, P. R. China
Tel: +86-717-6369520, 6369558
Fax: +86-717-6370680
Email: yefood@angelyeast. com




